MolAD
QSAR Model Applicability Domain Determination
Overview
This repository explores advanced methodologies for determining the applicability domain of a Quantitative Structure-Activity Relationship (QSAR) model. The approaches employed leverage sophisticated techniques such as Principal Component Analysis (PCA) and Tanimoto Similarity Matrix coupled with Multidimensional Scaling (MDS) algorithms.
Methodologies
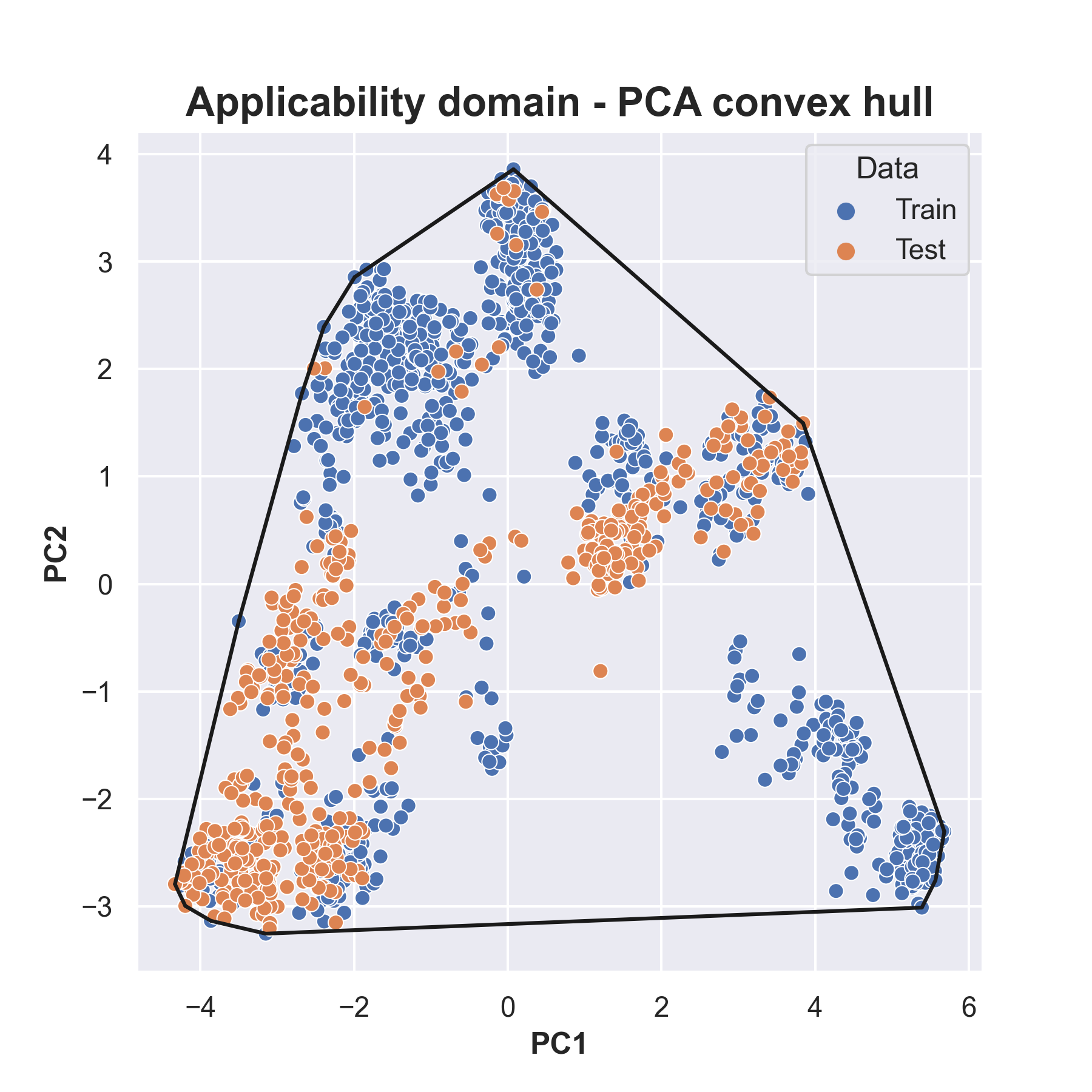
Approach 1: PCA-based Dimension Reduction and Convex Hull
In the first approach, molecular fingerprints undergo precise dimension reduction through Principal Component Analysis (PCA), resulting in a streamlined 2-dimensional representation. Subsequently, these observations are meticulously visualized on a plane, and a Convex Hull algorithm is applied to precisely delineate the confidence region.
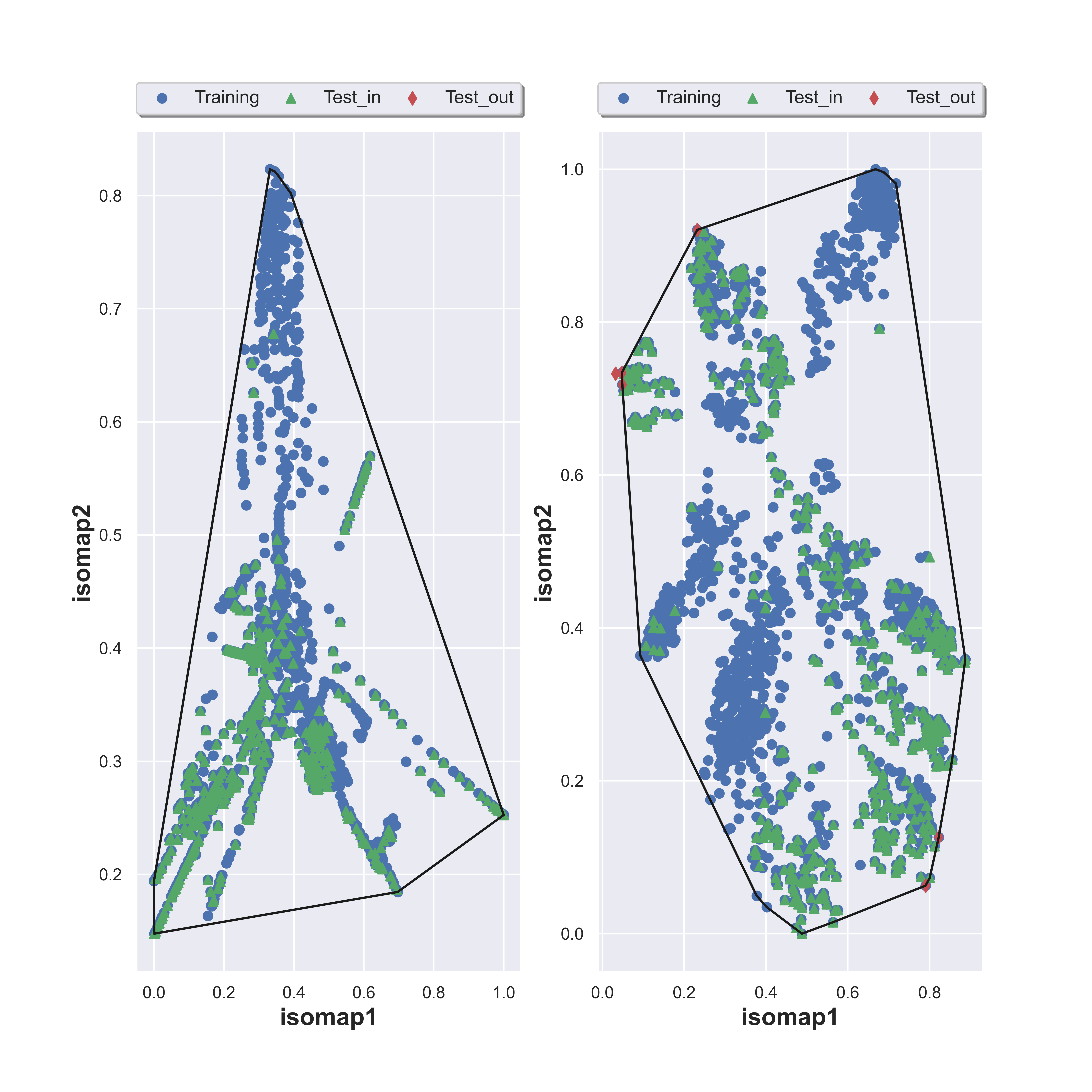
Approach 2: Tanimoto Similarity Matrix, MDS and Convex Hull
The second approach commences by calculating a Tanimoto similarity matrix, meticulously pairing molecules from both the training dataset (m molecules) and screening dataset (n molecules). This intricate process results in a (m+n) x (m+n) matrix. Following this, dimension reduction to 2 dimensions is applied to the matrix through state-of-the-art MDS algorithms. The resulting observations are meticulously plotted on a plane, and akin to the first approach, a Convex Hull algorithm is employed for precise delineation of the confident region.
Instructions
For comprehensive instructions and detailed code implementation, please refer to the Applicability_domain_screen.ipynb notebook.
Usage Guidelines
Follow the outlined steps in the notebook to effectively apply the proposed methodologies for determining the applicability domain of your QSAR model.
Contact
For any questions or inquiries, please contact The-Chuong TRINH.
Contributions
Contributions are highly encouraged. If you have valuable suggestions, improvements, or innovative ideas, please feel free to open an issue or submit a pull request.
Requirements
This module requires the following modules:
Installation
Clone this repository to use
Note
Updating…
Contributing
Please visit the MolAD